

 |

|
INDEX
- Suggested Use for StellaBase Search Engines
- Genome Comparison
- Gene Family Search
- Keyword Search for Pfam Family
Suggested Uses
This site may be used for manifold purposes. Primers, genetic stock,, and gene expression patterns may be retrieved using very straight forward query forms. Links within these pages are also available which allow users to upload data.
Users may find genes of interest through either a sequence homology search function (StellaBlast) or a keyword search function to locate all genes of interest which have been shown to map to a specific gene family motif within a user-specified E-value criterion. Once a sequence of interest has been identified, detailed information regarding that sequence may be retrieved using a gene search function. Links are present to allow users to submit comments about these sequences.
Additionally, interspecific genomic comparisons are possible. Users may compare gene content of a particular gene family between lineages using gene family search. Users may also search for gene families which are present and absent in user defined lineages using a genome comparison search function. Additional information regarding this search is available below.
Gene Family Search
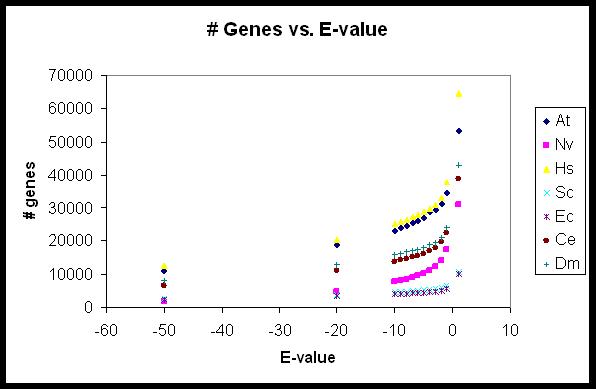
In the search for gene families, you must enter either a Pfam accession number or a Pfam protein family name in the textbox and check the appropriate radio button. The E-value threshold which you choose will have a significant impact on your results, as shown in the figure below.
Keyword Search for Pfam Family
This search function looks for search terms within the description of a gene family as defined by Pfam. You should use miminal search terms initially and then narrow down your search by adding search terms.
For example, If you are searching for '2',5' RNA Ligase' you should start your search with simply 'Ligase' and then further refine your search if neccessary.
Genome Comparison
This tool compares the genomes of a number of organisms. It allows users to select organisms to be included in the search and look specifically for gene families that are either present or absent in each selected organism.
Two searches are available- users can either search for gene families that are present in any of the organisms selected present (uncheck check box) or search for gene families that are present in all of the organisms selected present (leave check box checked).
For example, if a user wishes to search for gene families that are present in deuterostomes (e.g., mouse and human) yet absent in protostomes (e.g., fly and worm), you would want to search for all gene families that are present in any deuterostome yet absent in protosomes. As such, you would uncheck the box labeled 'Return only those proteins which are present in ALL of the species selected Present.'
If however, a user wishes to search for gene families that are present in both deuterostomes and cnidarians (e.g., Nematostella) yet absent in protosotomes, a user would want only those genes returned that are in both the deuterostomes and the cnidarians. As such, users would leave the box checked to make sure that only gene families are returned that are at the intersection of both deuterostomes and cnidarians yet are absent in protostomes
Questions, comments, suggestions: busully@bu.edu